what should you study if you want to know which genes are expressed by cells?
Fifty-fifty though about every cell in an organism's body contains the same set of genes, only a fraction of these genes are used in any given cell at any given time. It is this carefully controlled design of what is chosen "cistron expression" that makes a liver cell different from a muscle cell, and a good for you cell different from a cancer cell. But how can researchers decide which genes are "turned on" and when?
How do scientists measure cistron expression?

Effigy 1: Siamese cats have colored \"points\" because of a temperature-sensitive pigmentation gene. In cooler areas of a cat'south body (nose and paws), this factor is expressed to a greater degree.
Gene expression is dynamic, and the aforementioned gene may act in different means under different circumstances. For example, imagine that two organisms take similar genotypes merely unlike phenotypes. What is the cause of this variation in phenotype? Could the difference stalk from differing regulation of gene expression? Could temperature affect expression of the organism'south Deoxyribonucleic acid (Figure 1)? Might some other factor be responsible? To go well-nigh answering these types of questions, researchers often use laboratory techniques such as a Northern blot or serial analysis of gene expression (SAGE). Both of these techniques make it possible to place which genes are turned on and which are turned off within cells. Afterward, this data can be used to assist decide what circumstances trigger expression of various genes.
Both Northern blots and SAGE analyses work past measuring levels of mRNA, the intermediary between Deoxyribonucleic acid and protein. Remember, in society to activate a gene, a cell must first copy the Dna sequence of that cistron into a piece of mRNA known every bit a transcript. Thus, by determining which mRNA transcripts are present in a cell, scientists can determine which genes are expressed in that cell at different stages of development and under different environmental conditions.
Northern blots: What are they, and how do they work?
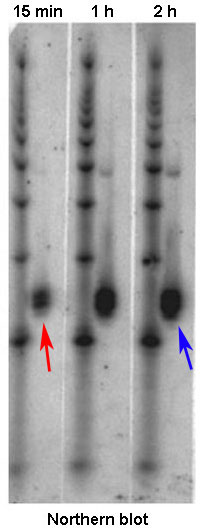
Effigy 2: A Northern blot allows comparing of mRNA levels, which are a straight reflection of gene expression. This blot in a higher place shows the tracking of mRNA levels in a jail cell sample at various intervals later exposure to a drug. At fifteen minutes after exposure, mRNA levels are low (red arrow, pocket-size black mark); at two hours after exposure, these levels have increased (bluish arrow, larger black mark).
The quantity of mRNA transcript for a single factor directly reflects how much transcription of that gene has occurred. Tracking of that quantity will therefore indicate how vigorously a gene is transcribed, or expressed. To visualize differences in the quantity of mRNA produced by different groups of cells or at different times, researchers oftentimes use the method known as a Northern blot. For this method, researchers must outset isolate mRNA from a biological sample by exposing the cells within it to a protease, which is an enzyme that breaks down cell membranes and releases the genetic material in the cells. Adjacent, the mRNA is separated from the Dna, proteins, lipids, and other cellular contents. The different fragments of mRNA are then separated from 1 another via gel electrophoresis (a technique that separates molecules by passing an electrical current through a gel medium containing the molecules) and transferred to a filter or other solid support using a technique known as blotting. To identify the mRNA transcripts produced by a particular gene, the researchers next incubate the sample with a short piece of unmarried-stranded RNA or DNA (also known every bit a probe) that is labeled with a radioactive molecule. Designed to be complementary to mRNA from the factor of involvement, the probe will bind to this sequence. Afterward, when the filter is placed against X-ray moving picture, the radioactivity in the probe volition betrayal the film, thereby making marks on it. The intensity of the resulting marks, called bands, tells researchers how much mRNA was in the sample, which is a straight indicator of how strongly the gene of involvement is expressed (Figure 2).
Is information technology possible to report the expression of multiple genes simultaneously?
Until recently, scientists studied gene expression by looking at merely one or very few factor transcripts at a fourth dimension. Thankfully, new techniques now brand large-scale studies of cistron expression possible. 1 such technique is SAGE (serial analysis of cistron expression). A method for measuring the expression patterns of many genes at once, SAGE not merely allows scientists to analyze thousands of gene transcripts simultaneously, but it also enables them to determine which genes are active in different tissues or at different stages of cellular development.
How does SAGE work?
SAGE identifies and counts the mRNA transcripts in a jail cell with the help of short snippets of the genetic lawmaking, called tags. These tags, which are a maximum of 14 nucleotides long, enable researchers to match an mRNA transcript with the specific gene that produced it. In most cases, each tag contains enough information to uniquely identify a transcript. The proper noun "serial analysis" refers to the fact that tags are read sequentially every bit a continuous string of data.
The basic steps of the SAGE technique are outlined beneath.
Capturing mRNA
To begin a SAGE analysis, researchers must first separate the mRNA in a sample from the other cellular contents. To do this, they attach long strips of thymine nucleotides to tiny magnetic chaplet. When researchers flush the contents of a cell over the chaplet, these thymine strips form complementary base of operations pairs with the poly-A tails of the mRNA molecules. Thus, when the flushing process is complete, the mRNA transcripts from the sample are captured because they are attached to the magnetic beads, while the other contents of the cells flush by the beads and are discarded.
Rewriting mRNA into cDNA

Figure 3: Reverse transcription converts mRNA into cDNA.
mRNA is more fragile than Dna, which makes it difficult to handle and analyze. To solve this problem, researchers oft convert mRNA samples into complementary Deoxyribonucleic acid sequences, or cDNA. This is washed past reversing the natural process a cell uses to brand mRNA from DNA, a method known every bit contrary transcription. The reverse transcription process doesn't utilize DNA polymerase or RNA polymerase; instead, it employs a special enzyme called reverse transcriptase. This enzyme makes cDNA sequences that are complementary to each mRNA transcript, substantially creating a converted form of the aforementioned sequence (Figure 3). This new unmarried-stranded cDNA is then converted into a double-stranded cDNA molecule.
Cutting tags from each cDNA
To begin the adjacent portion of SAGE, the researchers use a cutting enzyme to piece off short segments of nucleotides, called tags, at designated positions in each cDNA molecule. Next, two tags from each cDNA are combined into a unmarried unit. These tags and so go the representative for the gene they came from, and they human action equally a unique identifier in the form of a stand-in. Without having to process the entire mRNA sequence thereafter, scientists can utilize these shorter tag sequences to keep runway of whether a specific gene was expressed in mRNA form.
Linking tags together in chains for sequencing
Later on the dissimilar tags have been made from each mRNA sequence, they are next linked together into long chains called concatemers. These concatemers therefore contain representatives of mRNAs from a grouping of genes. Linking the tags together in a concatemer is important, because it ways that researchers will later exist able to read thousands of tags at once during the assay portion of the SAGE process.
Copying and reading the chains
Although the researchers now possess concatemers representing the genes expressed in the sample, they need multiple copies of these concatemers if they wish to run the molecules through a sequencing motorcar. Thus, only before sequencing, the concatemers are inserted into bacteria, and through their own replication procedure these bacteria make millions of copies of each concatemer concatenation. This stride increases the volume of cloth, and it therefore ensures that there is a baseline amount of material necessary for a sequencing machine. Later that, researchers use a sequencing machine to decode and read the long string of nucleotides in each concatenation.
Identifying and counting the tags
Finally, a computer processes the data from the sequencing machine and compiles a listing of tags. By comparing the tags to a sequence database, the researchers tin can identify the mRNA (and ultimately the gene) that each tag came from. By later on counting the number of times each tag is observed, the researchers can besides estimate the degree to which a detail gene is expressed: the more ofttimes a tag appears, the greater the level of factor expression.
What can researchers learn from SAGE?
Compared to other techniques for measuring gene expression, SAGE offers a significant reward because it measures the expression of both known and unknown genes. Sometimes, when analyzing SAGE data, computers cannot find matches for sure tags in their sequence databases. What does this hateful? Interestingly, a lack of matches indicates that the mRNA used to produce these tags is associated with genes that take not been studied earlier. In this mode, SAGE has been used to discover new genes involved in a diverseness of diseases.
Are there other ways to measure gene expression?
More about measuring gene expression
In addition to Northern absorb tests and SAGE analyses, at that place are several other techniques for analyzing factor expression. Nearly of these techniques, including microarray assay and reverse transcription polymerase chain reaction (RT-PCR), work by measuring mRNA levels. However, researchers can also analyze factor expression by directly measuring protein levels with a technique known as a Western blot.
Source: https://www.nature.com/scitable/topicpage/gene-expression-is-analyzed-by-tracking-rna-6525038/
0 Response to "what should you study if you want to know which genes are expressed by cells?"
Post a Comment